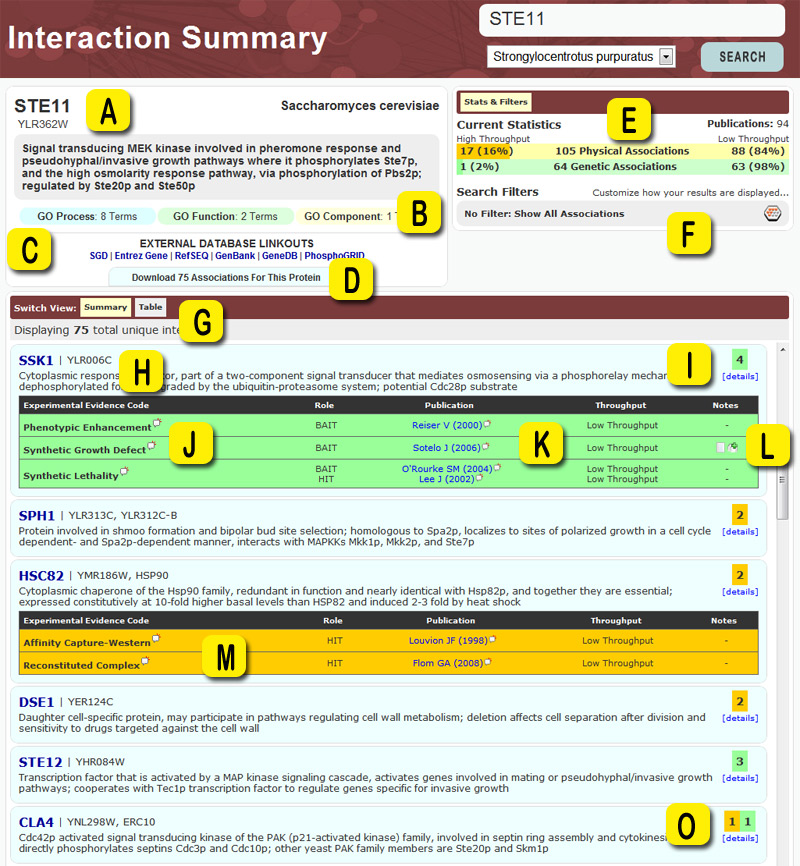
BioGRID Search Results Layout
BioGRID Search Results Layout
The following is an example layout viewed when searching for your protein of interest within the BioGRID. We've outlined all areas of interest with yellow letters, and defined what each area represents in the definitions at the bottom of the page.
Layout Legend
- A - Standard annotation data for search results including official name, synonyms, organism name, and description.
- B - Gene Ontology summary for the search result. If you're interested in seeing the full details for this result you can click on each category to expand the GO Summary into a list of mapped GO Entries.
- C - External links to 3rd party databases. These links generally point to model organism databases like SGD, MGI, FlyBase, etc. and also large genome resources such as Entrez-Gene and Ensembl.
- D - Click here to download a dataset consisting only of interactions listed on this search result page.
- E - A quick summary of the number of interactions represented in the current results. In this case there are 105 physical associations and 64 genetic associations listed. The colored bar behind each number denotes the relationship between high throughput and low throughput interactions. In this case, 16% of the physical associations are HTP and 2% of the genetic associations are HTP.
- F - By clicking here, you can activate or deactivate a filter for the interactions shown. Filters allow you to customize which results are featured prominently on searches.
- G - This list of buttons allows you to switch the current view and look at the data in an alternate format.
- H - Annotation data for each of the resulting interactors with your search result. As above, the interface provides official names, aliases, and descriptions. Each interactor is hyperlinked to allow for circular searching. Simply click on the name and you will automatically pull up all curated interactions involving that particular gene, without having to redo your keyword searching.
- I - Quick summary showing that this record (SSK1) has supporting evidence from 4 different associations. The green color represents that all 4 are genetic. You can mouse over this box to get more details or click it to get a summary listing directly below it. In the 'Summary' view interactors are ordered acccording to the following scheme:Protein interactions are weighted greater than genetic interactions (1.5 vs. 1.0). Any interactions you have filtered out are given a weighting of 0.01. So if STE11 has 10 protein and 10 genetic interactions it gets a score of 25. The interactors are then ordered by score descending.
- J - Summary table showing grouped interactions and their associated annotation. You can click on the white speech bubbles behind each experimental evidence code to see a description of that code. This table is initially not show, but is displayed when clicking the summary box shown in item I.
- K - Publication supporting this association. You can click on the speech bubble to display more details about the publication or click on the author name to go to a separate page where you can download all interactions associated with that publication. Publications show the first authors last name and the date in which the publication was published.
- L - If an association has additional notes, such as phenotypes, qualifications, modifications, quantitative scores, etc, you will see small icons in the notes column. Simply click on the icons to see a detailed listing of the information available.
- M - If an association is represented by physical experimental evidence codes, the associated boxes will be orange in color.
- O - If an interactor has both physical and genetic experimental evidence codes, the summary will display multiple boxes showing each different type separately.